A Mathematical Approach to Countering Resistant Bacteria
Here’s an example of some of the fascinating research being done today at the interface of mathematics and biology. The two fields have become ever more intertwined as our knowledge of genetics has deepened. The subtle and not so subtle interactions between microbes and their host, the multitudes of genes that give rise to behavior, the modeling of metabolic pathways, large-scale sequencing projects—all are stubbornly complex to puzzle out, and all bend more easily with the aid of innovative statistical techniques and advances in computing power.
Among the more pressing topics in medicine today concerns whether antibiotics have outlived their usefulness or whether we simply need to rethink how we use them. When German pathologist Gerhard Domagk discovered the antimicrobial effects of Prontosil in the 1930s, it was heralded as one of the greatest therapeutic advances in medical history. Yet even then, scientists were wary of the long-term drawbacks of their overuse. Almroth Wright predicted that bacteria would evolve resistance to these agents before we observed the phenomenon directly, while Nobel laureate Alexander Fleming (of penicillin fame) cautioned that they should be administered only in specific circumstances and in the proper dosage so as to minimize the risk of resistant pathogens.
While these words of warning have proved especially prescient in the wake of our current crisis, we are not entirely to blame. Bacteria, fungi and algae have been locked in a campaign of chemical warfare for more than a billion years. Like Darwinian selection itself, antibiotics and resistant genes long predate humanity; we merely gave them names. Microbes, moreover, outnumber us billions to one, with much greater genetic diversity and shorter reproductive cycles. This allows mutations to spread through populations quickly and gain the upper hand before new pharmaceutical trials can even get off the ground. Lastly, bacteria also are fond of passing helpful genes around to their neighbors through a process called horizontal gene transfer, and the immense diversity of the microbial jungle means there will always be resistant DNA close at hand.
As Martin Blaser writes in his latest book, Missing Microbes:
“Another implication of the nature of resistance is that there will be no easy solution to the problem. We will never make resistance go away, because Darwin was correct in his theories. There always will be strong selection for resistance when populations encounter stress, in this case, microbes under antibiotic pressure. A corollary is that we will never invent a superantibiotic that cures everything. Microbes are too diverse, and Nature will always come up with new ammunition.” (pp. 80-81)
Given the natural processes which govern life on earth, the crossroads at which we now find ourselves was perhaps a biological inevitability. Evolution is an arms race fought with genetics across time. Rewrites that give contestants a replicative edge reconfigure the battle lines; old strategies wane in strength. While most bacteria are beneficial, some, like Streptococcus and Salmonella, are pathogenic, interacting with host physiology in harmful ways. To counter them, we have antibiotics, which work well enough for wiping out particularly devastating ills but effectively wage genocide on our microbiome, killing off both good and bad residents and upsetting gut ecology. Not only can these collateral damage “grenades” lead to unintended complications down the road, evolution does not sit still. Any mutations that limit antibacterial efficacy quickly outcompete those that don’t, in the process generating more resilient microbes for us to outwit.
The doctor-patient relationship, along with common agricultural practices, have certainly not helped matters, however. For decades we have over-prescribed these broad-spectrum solutions for misdiagnosed and nonbacterial infections, relied on them as “just in case” remedies when less invasive ones would do, and pumped them into the animals we farm and the food we consume. This escalating confluence of dependence on antimicrobials and our commitment to an antiseptic lifestyle has served to fast-track evolution in the form of resistance.
Many immunologists have suggested we are moving into the “post-antibiotic” era, but what if we could predict the evolutionary trajectory of certain bacteria when subjected to antibiotics? Might we be able to substitute new drugs before resistance emerges and spreads? That’s the idea behind new research pioneered by Miriam Barlow from University of California and published in PLoS ONE this month. Together with mathematicians from American University in Washington, D.C., they were able to apply a probability analysis to bacterial evolution.
Rewinding the Genetic Clock
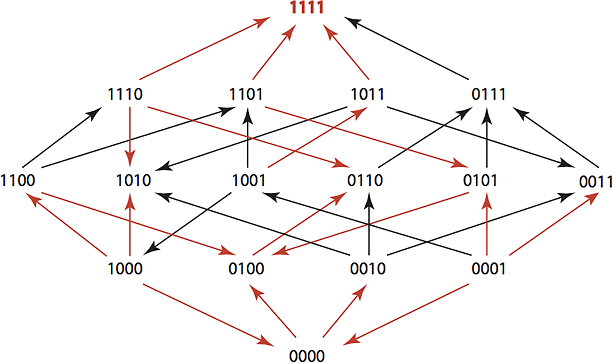
The researchers exposed E. coli to 15 different antibiotics, taking note of the differences to TEM-1, a protein involved in 90% of penicillin-derived drugs. A wild type TEM-1—which can be conceptualized as the “standard” form of the protein, or one unexposed to antibiotics—confers resistance to penicillin only, while its variants (other genotypes) tend to expand resistance to other classes of antibiotics such as cephalosporins.
The team’s goal was to see how consistently they could revert a population of TEM-1 genotypes to wild type status, thereby narrowing the range of resistance. Since nearly all known TEM-1 variants differ by no more than four mutations, the team used a four-digit model to map out the changes in amino acids. For example, the wild type TEM-1 was assigned a label of “0000”, and each mutation was represented by a “1” in the location where the substitution occurred.
Using specialized software, Barlow and her team were able to devise treatment plans that could return TEM-1 descendants to their wild type state 60% of the time, a significant improvement over the antibiotic cycling schemes in place today. While the growth rates varied among genotypes, only a short list of antibiotics were required to accomplish the feat in each case. By playing pilot to evolution in this way, we could theoretically drive resistant bacteria in a specified direction and then switch to a drug effective against the genotype in question. They dubbed their software suite the “Time Machine” for its ability to regress resistance genotypes to ones “that existed at some prior point in time”.
Though these results are encouraging, it is not clear how clinically relevant they are. A lab setting in which tight constraints are maintained throughout is one thing; a crowded hospital overflowing with bacterial and antibacterial diversity is another. Microbes living in communion with other microbes tend to share genetic defenses, a feature not available to those grown in isolation. Moreover, different host environments supply a wider playspace for natural selection to act, possibly serving up a greater variety of genotypes than those captured by the study. And while such algorithmic exploitation could theoretically aid us in our fight against resistance, we would still need a better idea of how long each drug should be in rotation and the recommended dosages necessary to complete the wild type selection process.
That under certain conditions bacteria nestle along statistically predictable alleyways is an interesting result in and of itself, but more research in the form of field tests and clinical trials will be required to comment on its real-world validity.
“At the heart of what everybody wants to know is how predictable is evolution—and if it’s predictable, can we reverse it?” he says. “It’s really hard, but you’ve got to try something.”
Further reading:


Comments